From a simple statistical perspective, fMRI data can be considered as a large number of individual voxel time series. If these individual times series were independent, one could use any of the models from traditional time series analysis. For example, one could treat the brain as a linear system modulating the stimulus as its input. A model for this system would simply estimate the coefficients of the transfer function. Of course, there are other inputs such as heartbeat and respiration as well as other sources of data interference which would also need to be included. Thus, an ideal model would consist of an equation for each voxel that accounts for the true brain signal as well as all sources of systematic and random noise. By estimating all sources of noise and removing them appropriately for each voxel, an accurate estimate of brain activity could be found.
As can be imagined, an ideal model for fMRI data activation is highly
impractical. A typical fMRI experiment may consist of
![]() voxels; therefore, a model that consists of a separate
equation for each voxel would be quite cumbersome. Furthermore, the
mere identification of each and every noise source in an fMRI
experiment alone is a difficult task. Thereby a precise quantification
of the effects of each noise source would be nearly impossible. In
order to model and analyze fMRI data in a practical manner,
researchers often take an approach that is similar to that of solving
any giant problem; that is, by breaking it down into piece-wise,
practical steps. This approach allows for easier understanding of each
step that is carried out (since they are performed one at a time)
while also making the analysis computationally manageable. The
piece-wise analysis steps involve first modeling and removing
identifiable noise sources, then statistically evaluating the
corrected data to estimate the amount of brain activation.
voxels; therefore, a model that consists of a separate
equation for each voxel would be quite cumbersome. Furthermore, the
mere identification of each and every noise source in an fMRI
experiment alone is a difficult task. Thereby a precise quantification
of the effects of each noise source would be nearly impossible. In
order to model and analyze fMRI data in a practical manner,
researchers often take an approach that is similar to that of solving
any giant problem; that is, by breaking it down into piece-wise,
practical steps. This approach allows for easier understanding of each
step that is carried out (since they are performed one at a time)
while also making the analysis computationally manageable. The
piece-wise analysis steps involve first modeling and removing
identifiable noise sources, then statistically evaluating the
corrected data to estimate the amount of brain activation.
Basically, two approaches can be employed to correct for known sources of bias and variance in fMRI data. These are correction of the data at the time of collection (proprocessing) and correction after data collection (post-processing). We use both approaches, often in tandem. For example, we use various forms of head restraint to (partially) eliminate head motion (proprocessing) and in addition we use post-processing to correct the data for head motion in addition.
We now give several examples of how data correction can be done in
a post-processing manner. Some of these examples outline how many
fMRI data processing steps are currently carried out using FIASCO
(Functional Image Analysis Software - Computational
Olio), which is a software collection that has been
developed by the authors of this paper together with others and is
currently used by numerous groups who analyze fMRI data. Details can
be found at http://www.stat.cmu.edu![]() fiasco.
fiasco.
These examples also demonstrate that large amounts of computation are often needed to remove bias and variance in fMRI data.
Noise from the equipment will first be addressed. Baseline noise or DC-shift noise can be corrected fairly easily, as it is a well understood source of noise in the data. To adjust for baseline noise, the idea that the k-space data should (approximately) be oscillating around 0 at the highest spatial frequencies is taken into account. If this is not the case in the true data, the mean value at which the high frequency data is oscillating is computed, and the k-space data is shifted by a constant prior to correct for this source of noise (see [7]).
Nyquist ghosts are also well understood and can
therefore be corrected fairly easily. These ghosts arise from small
mistimings in the gradients with respect to the data collection. To
correct for these ghosts, a phase shift is applied to each line (the
same shift for each line in the ![]() -direction) of the complex-valued
k-space data in order to move the data back into its correct
location. The best phase shift for correction is estimated from the
data by finding a value which minimizes the magnitude of the ghosts.
Typically, the center portion
of the top and bottom few lines is chosen as the target
(see [7]).
-direction) of the complex-valued
k-space data in order to move the data back into its correct
location. The best phase shift for correction is estimated from the
data by finding a value which minimizes the magnitude of the ghosts.
Typically, the center portion
of the top and bottom few lines is chosen as the target
(see [7]).
Magnetic field inhomogeneities may or may not vary with time. Those which do not vary with time can be corrected by ''shimming'' the magnet. Small magnetic objects are placed around the magnet in such a way as to reduce the inhomogeneity. Inhomogeneities associated with the subject being scanned can be corrected by dynamic shimming of the field. This is a procedure performed by the technologist at the time of the experiment. For further details see, e.g., [25] or [11].
To reduce the effects of magnetic field distortions in a post-processing manner, the following procedure can be carried out. First a phase map can be computed from the image information in the fMRI data. This map is thought to give an estimate of regions of inhomogeneities in the data. Next, a 2-D polynomial can be fit to the field map, which is then converted into a pixel shift map. The shifted pixels are moved back to their proper locations in order to correct for the magnetic field inhomogeneities; further details may be found in [26].
Noise from the experimental subject should also be corrected to improve data quality. As mentioned in the previous section, head motion is major problem in fMRI, since even relatively small amounts of head motion can lead to false positive and false negative regions of activation. Many techniques have been considered to reduce the amount of head motion in fMRI data. These (again) can be classified into the categories of proprocessing and post-processing.
To reduce the amount of head motion that occurs at the time of the fMRI scan, external restraining devices are often used. These devices range from pillows and straps to dental bite bars and even thermoplastic face masks; see [20]. Head restraints can greatly reduce the amount of head motion but unfortunately cannot alleviate head motion (or at least brain motion) altogether; see [14]. In fact, [53] have suggested that some types of restraints can paradoxically increase head motion because of the discomfort they cause at pressure points. Furthermore, some types of restraints may not be appropriate for certain subject types such as small children and mentally disturbed subjects.
Also experimental design can be used to reduce the amount of apparent head motion. Often head motion is associated with presentation of a stimulus or with physical response to the stimulus. Careful experimental design can eliminate or largely alleviate such effects.
A second way to reduce the effect of head motion at the time of the
scan is through the use of navigator echos. Navigator echos are used
before slice acquisition in order to detect displacements of the head.
These displacements are then used to adjust the plane of excitation of
the collected image accordingly. Examples include the use of
navigator echos to correct for displacements in the ![]() -direction
(see [32]),
and the use of navigator echos to correct for
inter-image head rotation (see [33]).
-direction
(see [32]),
and the use of navigator echos to correct for
inter-image head rotation (see [33]).
A final example of a prospective method to reduce head motion is a visual feedback system that was developed by [46]. This system provides a subject with information about their head location through a visual feedback system. By using this device the subject can tell if their head has moved during the scan and can immediately correct for it.
Proprocessing techniques have many benefits. For example, the collected fMRI data presumably has less head motion than it would have without the adaptation to reduce for head motion. Therefore, there is less need for post-processing of the data and less need to resample or interpolate the fMRI data; interpolation of fMRI data can introduce additional errors. On the other hand, these techniques often require specialized collection sequences or hardware, which can further necessitate specialized analysis software. These techniques can also lead to the need for longer data collection times.
Post-processing techniques are often more feasible for fMRI researchers because they do not require specialized collection sequences or hardware. Post-processing is mainly carried out through image registration, which involves two main steps. The first is estimation of how much motion has occurred in the collected data, and the second is correction of the estimated motion through resampling (or interpolation). This process can be carried out in two dimensions for estimation and correction of in-plane motion or three dimensions for whole head motion.
To estimate how much head motion has occurred in an fMRI data set, a reference image must first be chosen. Any image can be chosen for this purpose (typically, the first or middle image in the series), but a composite image such as the mean can also be used. Next, each image in the series is compared to the reference image using a chosen feature of the images. Image intensity is often the chosen feature, but anatomical landmarks can also be used. Mathematically, the comparison process finds the geometrical shift required between the reference image and the target image to minimize an optimization criteria. The shift that minimizes this criteria is considered to be the amount of motion that has occurred.
For two dimensional rigid motion between the current image and the
reference image we consider translations in ![]() and
and ![]() , and
in-plane rotations of
, and
in-plane rotations of ![]() . In three dimensions rigid motion is
even more complicated because translations can occur in
. In three dimensions rigid motion is
even more complicated because translations can occur in ![]() ,
, ![]() , and
, and
![]() , and rotations can occur in the
, and rotations can occur in the ![]() ,
, ![]() , or
, or ![]() directions. Criteria to be minimized between the reference image and
the current image have included weighted mean square error
(see [8]).
variance of the image ratios
([51]),
and mutual information
([27]).
directions. Criteria to be minimized between the reference image and
the current image have included weighted mean square error
(see [8]).
variance of the image ratios
([51]),
and mutual information
([27]).
Once the motion estimates are computed, the data are moved back to their appropriate locations. This is carried out by resampling or interpolation. If data relocation is not performed in the best possible way, false correlations between image voxels can be introduced. [8] developed a Fourier interpolation method to prevent introduction of these errors. Fourier interpolation is based on the Fourier shift theorem and uses the fact that a phase shift in k-space is equal to a voxel shift in image space. By this property, translations are corrected using phase shifts, and analogously rotations are corrected by k-space rotations. Rotations in k-space are implemented by factoring the two-dimensional rotation into a product of three shearing matrices. A two-dimensional rotation matrix
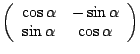 |
![$\displaystyle \notag \left(\begin{array}{cc} 1 & -\tan\displaystyle\frac{\alpha...
...ay}{cc} 1 & -\tan\displaystyle\frac{\alpha}{2}\\ [2mm] 0 & 1 \end{array}\right)$](img8239.gif) |
Once the rotations are represented by three shearing steps, these too
can be corrected using phase shifts in Fourier space; for details see
[8].
In a comparison of different types of fMRI
data interpolation algorithms, [10] found that
full Fourier interpolation was the only method which completely
preserved the original data properties. Table 4.1 is taken from that
paper and shows the error introduced by each method in performing the
following motion. Each image was rotated by ![]() radians,
translated
radians,
translated ![]() pixel on the diagonal, translated back
pixel on the diagonal, translated back ![]() pixel, and
rotated back
pixel, and
rotated back ![]() radians. Many of the algorithms had major errors
at the edges of the image so the statistics were only computed over
a central portion where no edge effects occurred.
radians. Many of the algorithms had major errors
at the edges of the image so the statistics were only computed over
a central portion where no edge effects occurred.
| Method | MSD |
| Fourier | |
| WS16 | |
| WS8 | |
| WS4 | |
| NN | |
| Quintic | |
| Cubic |
|
| WS2 |
|
| Linear |
|
Although three dimensional head motion seems more realistic for correction of subject movements in fMRI research, the process of estimating the six motion parameters that jointly minimize the optimization criteria can be computationally expensive. Also note that these six parameters only account for rigid body head motion. Data is actually collected one slice at a time; each slice is collected at a slightly different time. Consequently, there is no three-dimensional image of the head which can be used as a target for three dimensional registration. In spite of these considerations, the use of post-processing motion correction techniques does not necessitate specialized collection equipment, and if done correctly, can be quite accurate for small motions of the head. Large head movements, greater than say a millimeter or two, usually affect the data so much that it cannot reasonably be used.
![\includegraphics[width=\textwidth,clip]{text/4-4/fig6.eps}](img8249.gif)
|
A second significant noise source from the experimental subject is physiological noise, which is mainly attributed to the subject's heart beat and respiration. Many methods have been introduced to reduce the noise and variability caused by physiological noise. These again have included techniques that address the problem in a proprocessing manner, and techniques that reduce this noise during post-processing of the data by various modeling and subtraction techniques ([2,5,24,19]).
Acquisition gating is most commonly used for the collection of cardiac MRI data, but it can also be used for fMRI. This proprocessing technique only collects the MRI data at certain points of the cardiac cycle so that noise effects due to cycle variations can be reduced ([21]). Although useful for cardiac MRI, it is not as practical for fMRI research because it does not allow for continuous and rapid data collection, which is usually desirable for fMRI research.
Post-processing techniques to correct for physiological noise have included retrospective image sorting according to the phase of the cardiac cycle ([6]), phase correction through the assumption that the phase is uniform for each point in k-space over many images ([52]), and digital filtering ([2]). Each of these techniques have certain benefits in physiological noise correction; however, each can also compromise the fMRI data. For example, image reordering results in loss of temporal resolution, and digital filtering requires the ability to acquire images rapidly as compared to the physiological noise signals.
Other approaches to physiological noise correction have included modeling of the physiological data signals in the fMRI data and subtracting out their effects. For example, Hu modeled respiration with a truncated Fourier Series and subtracted this curve from the magnitude and phase components of the k-space data ([24]). [19] carried out a similar Fourier modeling and subtraction procedure in image space. Alternatively, Mitra and Pesaran modeled cardiac and respiratory effects as slow amplitude, frequency-modulated sinusoids and have removed these components from an fMRI principal component time series, which was obtained using a spatial frequency singular value decomposition [38].
In a more recent study carried out by the authors, cardiac data and respiratory data were found to be significantly correlated with the k-space fMRI data. These correlations are a function of both time and spatial frequency ([36]). Because of these correlations, the physiological effects can simply be reduced by first collecting the physiological data along with the fMRI data, regressing the fMRI data onto the physiological data (with the appropriate temporal considerations), and subtracting out the fitted effects ([37]). Figure 4.6 shows the cross-correlations between the k-space phase at one spatial frequency point and the cardiac data for one experiment.
In addition to direct modeling of known sources of noise, we also attempt to reduce the effect of unknown sources of noise. For example, we routinely replace outliers with less extreme values. As another example we currently remove a linear temporal trend from each voxel times series, although we have no explanation for the source of this linear trend.
The very first fMRI experiments claimed to have discovered brain activation based on simply taking the difference between the average image in one experimental condition from the average in an alternate experimental condition. There was no notion of variability. It quickly became apparent that accounting for the variability was very important and fMRI analyses started using t-tests, independently on each voxel. It then became clear that multiple testing was a very serious problem; we will discuss this in more detail below. The t-test (and for experiments with more than two levels, F-test) became the standard analysis method. However, it is clear that although the stimulus switches instantaneously between experimental conditions, the brain response (presumably being continuous) must transition to the new state and there will be some time lag before the transition is complete. This has led to a variety of ad hoc ''models'' for the shape and lag of this transition (Gamma functions, Poissons, etc.) (see, for example, [30]).
There has been some work developing reasonable non-parametric models for the voxel time courses ([16]) and there have been a number of ''time series'' modeling approaches, using spectral analysis ([30]) and AR models ([23]).
Because a typical brain image contains more than ![]() voxels, it
is clear that choosing a significance level of
voxels, it
is clear that choosing a significance level of ![]() will, even under
the null hypothesis of no difference in brain activity, lead to
will, even under
the null hypothesis of no difference in brain activity, lead to ![]() or more voxels being declared active. The earliest attempt to deal
with this problem in fMRI was the split t-test, wherein the data
were divided into two (or more) temporal subsets. T-tests were
calculated separately within each subset and a voxel was declared
active only if it was active in all subsets ([43]).
This certainly reduced the number
of false positives, but obviously caused considerable loss of power.
or more voxels being declared active. The earliest attempt to deal
with this problem in fMRI was the split t-test, wherein the data
were divided into two (or more) temporal subsets. T-tests were
calculated separately within each subset and a voxel was declared
active only if it was active in all subsets ([43]).
This certainly reduced the number
of false positives, but obviously caused considerable loss of power.
Researchers quickly converged on the Bonferonni correction where one simply divides the significance level by the number of tests as a safe way of dealing with the problem. The loss of power is huge and some researchers started developing methods to compensate. [13] proposed the contiguity threshold method, which relies on the presumption that if one voxel is active then adjacent voxels are likely to be active.
The false discovery rate (FDR) controlling procedure was then introduced as an alternate method for thresholding in the presence of multiple comparison testing. The FDR procedure allows the user to select the maximum tolerable FDR, and the procedures guarantees that on average the FDR will be no larger than this value. Details of the application of FDR to fMRI data can be found in [17].
As researchers in fMRI have discovered various general classes of statistical models, they have been applied to these data sets. They have had varying degrees of success and it is still the case that a linear model, often with both fixed and random effects, is the model of choice. It is reasonably well-understood and is not too far from what is actually believed. One critical problem is that as experimental designs have become more complex analyses have too; often researchers are fitting models blindly without fully understanding some of the underlying statistical issues. The more cautious return to their linear models.