One of the classical formulations of the two-group discriminant
analysis or the statistical pattern recognition problem involves
a mixture of two ![]() -dimensional normal distributions with a common
covariance matrix. The problem of two-group cluster analysis
with multiple continuous observations has also been formulated
in this way. Here, we have
-dimensional normal distributions with a common
covariance matrix. The problem of two-group cluster analysis
with multiple continuous observations has also been formulated
in this way. Here, we have ![]() independent observations
independent observations
![]()
![]() from the mixture density
from the mixture density
Consider the corresponding ''supervised learning problem'', where
observations on the random vector
![]() are
are
![]()
![]()
![]() . Here
. Here
![]() is an indicator variable which identifies the
is an indicator variable which identifies the ![]() th observation
as coming from the first
th observation
as coming from the first ![]() or the second
or the second ![]() component
component
![]() . The MLE problem is far simpler here with easy
closed-form MLE. The classificatory variable
. The MLE problem is far simpler here with easy
closed-form MLE. The classificatory variable ![]() could be called the
''missing variable'' and data
could be called the
''missing variable'' and data
![]() the missing data. The unsupervised learning problem could be
called the incomplete-data problem and the supervised learning
problem the complete-data problem. A relatively simple iterative
method for computing the MLE for the unsupervised problem could be
given exploiting the simplicity of the MLE for the supervised
problem. This is the essence of the EM algorithm.
the missing data. The unsupervised learning problem could be
called the incomplete-data problem and the supervised learning
problem the complete-data problem. A relatively simple iterative
method for computing the MLE for the unsupervised problem could be
given exploiting the simplicity of the MLE for the supervised
problem. This is the essence of the EM algorithm.
The complete-data log likelihood function for
![]() is given by
is given by
Now the EM algorithm for this problem starts with some initial
value
![]() for the parameters.
As
for the parameters.
As
![]() in (5.10) is a linear function of the
unobservable data
in (5.10) is a linear function of the
unobservable data
![]() for this problem, the calculation of
for this problem, the calculation of
![]() on the E-step is effected
simply by replacing
on the E-step is effected
simply by replacing ![]() by its current conditional expectation
given the observed data
by its current conditional expectation
given the observed data
![]() ,
which is the usual posterior probability of the
,
which is the usual posterior probability of the ![]() th
observation arising from component 2
th
observation arising from component 2
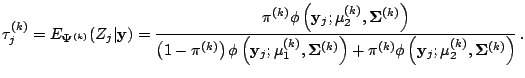 |
The M-step then consists of substituting these
![]() values
for
values
for ![]() in (5.11) to (5.13). The E- and M-steps
are then iterated until convergence. Unlike in the MLE for the supervised
problem, in the M-step of the unsupervised problem, the posterior
probabilities
in (5.11) to (5.13). The E- and M-steps
are then iterated until convergence. Unlike in the MLE for the supervised
problem, in the M-step of the unsupervised problem, the posterior
probabilities ![]() , which are between 0 and 1, are used. The
mean vectors
, which are between 0 and 1, are used. The
mean vectors
![]() and the covariance
matrix
and the covariance
matrix
![]() are computed using the
are computed using the
![]() as weights in weighted averages.
as weights in weighted averages.
It is easy to extend the above method to a mixture of
![]() multinormal mixtures or even to a mixture of
multinormal mixtures or even to a mixture of ![]() distributions from other (identifiable) families. For a detailed
discussion of the applications of the EM algorithm in the
resolution of finite mixtures and other issues of finite
mixtures, see McLachlan and Peel (2000).
distributions from other (identifiable) families. For a detailed
discussion of the applications of the EM algorithm in the
resolution of finite mixtures and other issues of finite
mixtures, see McLachlan and Peel (2000).
In survival or reliability analyses, the focus is the distribution
of time ![]() to the occurrence of some event that represents
failure (for computational methods in survival analysis see also
Chap. III.12). In many situations, there will
be individuals who do not
fail at the end of the study, or individuals who withdraw from the
study before it ends. Such observations are censored, as we know
only that their failure times are greater than particular values.
We let
to the occurrence of some event that represents
failure (for computational methods in survival analysis see also
Chap. III.12). In many situations, there will
be individuals who do not
fail at the end of the study, or individuals who withdraw from the
study before it ends. Such observations are censored, as we know
only that their failure times are greater than particular values.
We let
![]() denote the observed failure-time data, where
denote the observed failure-time data, where
![]() or
or ![]() according as the
according as the ![]() th observation
th observation ![]() is
censored or uncensored at
is
censored or uncensored at
![]() . That is, if
. That is, if
![]() is uncensored,
is uncensored, ![]() , whereas if
, whereas if ![]() , it is
censored at
, it is
censored at ![]() .
.
In the particular case where the p.d.f. for ![]() is exponential
with mean
is exponential
with mean ![]() , we have
, we have
The complete-data vector
![]() can be declared to be
can be declared to be
![]() where
where
![]() contains the unobservable realizations of the
contains the unobservable realizations of the ![]() censored random
variables. The complete-data log likelihood is given by
censored random
variables. The complete-data log likelihood is given by
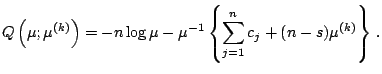 |
From (5.17), it can be seen that
![]() has the exponential family form (5.4) with
canonical parameter
has the exponential family form (5.4) with
canonical parameter ![]() and sufficient statistic
and sufficient statistic
![]() Hence, from (5.18), the E-step requires the calculation of
Hence, from (5.18), the E-step requires the calculation of
![]() .
The M-step then yields
.
The M-step then yields
![]() as the
value of
as the
value of ![]() that satisfies the equation
that satisfies the equation
Examples 1 and 2 may have given an
impression that the E-step consists in replacing the missing data
by their conditional expectations given the observed data at
current parameter values. Although in many examples this may be
the case as
![]() is a linear function of the missing
data
is a linear function of the missing
data
![]() , it is not quite so in general. Rather, as should be
clear from the general theory described in Sect. 5.2.1, the E-step
consists in replacing
, it is not quite so in general. Rather, as should be
clear from the general theory described in Sect. 5.2.1, the E-step
consists in replacing
![]() by its conditional
expectation given the observed data at current parameter values.
Flury and Zoppé (2000) give the following interesting example
to demonstrate the point that the E-step does not always consist
in plugging in ''estimates'' for missing data. This is also an
example where the E-step cannot be correctly executed at all
since the expected value of the complete-data log likelihood does
not exist, showing thereby that the EM algorithm is not
applicable to this problem, at least for this formulation of the
complete-data problem.
by its conditional
expectation given the observed data at current parameter values.
Flury and Zoppé (2000) give the following interesting example
to demonstrate the point that the E-step does not always consist
in plugging in ''estimates'' for missing data. This is also an
example where the E-step cannot be correctly executed at all
since the expected value of the complete-data log likelihood does
not exist, showing thereby that the EM algorithm is not
applicable to this problem, at least for this formulation of the
complete-data problem.
Let the lifetimes of electric light bulbs of a certain type have
a uniform distribution in the interval
![]() ,
,
![]() and unknown. A total of
and unknown. A total of ![]() bulbs are tested in two
independent experiments. The observed data
consist of
bulbs are tested in two
independent experiments. The observed data
consist of
![]() and
and
![]() , where
, where
![]() are exact lifetimes of a random sample of
are exact lifetimes of a random sample of ![]() bulbs and
bulbs and
![]() are indicator observations on a random sample of
are indicator observations on a random sample of ![]() bulbs,
taking value 1 if the bulb is still burning at a fixed
time point
bulbs,
taking value 1 if the bulb is still burning at a fixed
time point ![]() and 0 if it is expired. The missing data is
and 0 if it is expired. The missing data is
![]() . Let
. Let ![]() be the number of
be the number of
![]() 's with value 1 and
's with value 1 and
![]() .
.
In this example, the unknown parameter vector
![]() is a scalar,
being equal to
is a scalar,
being equal to ![]() .
Let us first work out the MLE of
.
Let us first work out the MLE of ![]() directly. The
likelihood is
directly. The
likelihood is
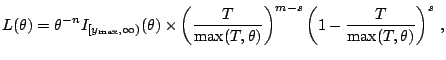 |
![$\displaystyle \notag \widehat{\theta} = \begin{cases}\tilde{\theta} & \mbox{if}...
...\geq 1\\ [-1mm] y_{\max} & \mbox{otherwise}\,. \end{cases}%\end{array} \right.
$](img1983.gif) |
Now let us try the EM algorithm for the case ![]() . The
complete data can be formulated as
. The
complete data can be formulated as
![]() and
the complete-data MLE is
and
the complete-data MLE is
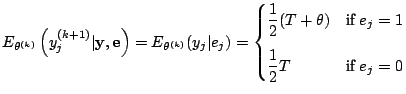 |
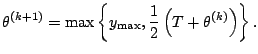 |
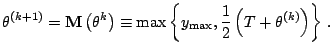 |
The reason for the apparent EM algorithm not resulting in the MLE
is that the E-step is wrong. In the E-step, we are supposed to
find the conditional expectation of
![]() given
given
![]() at current parameter values. Now given the data
with
at current parameter values. Now given the data
with ![]() , we have
, we have
![]() and hence the conditional
distributions of
and hence the conditional
distributions of ![]() are uniform in
are uniform in
![]() . Thus
for
. Thus
for
![]() the conditional density of missing
the conditional density of missing
![]() takes value 0 with positive probability and hence the
conditional expected value of the complete-data log likelihood we
are seeking does not exist.
takes value 0 with positive probability and hence the
conditional expected value of the complete-data log likelihood we
are seeking does not exist.
The EM algorithm will
converge very slowly if a poor choice of initial value
![]() were used.
Indeed, in some cases where the likelihood is unbounded on the edge
of the parameter space, the sequence of estimates
were used.
Indeed, in some cases where the likelihood is unbounded on the edge
of the parameter space, the sequence of estimates
![]() generated by the EM algorithm may diverge if
generated by the EM algorithm may diverge if
![]() is chosen too close to the boundary. Also, with
applications where the likelihood equation has multiple roots
corresponding to local maxima, the EM algorithm should be applied
from a wide choice of starting values in any search for all local
maxima. A variation of the EM algorithm (Wright and Kennedy, 2000)
uses interval
analysis methods to locate multiple stationary points of a log
likelihood within any designated region of the parameter space.
is chosen too close to the boundary. Also, with
applications where the likelihood equation has multiple roots
corresponding to local maxima, the EM algorithm should be applied
from a wide choice of starting values in any search for all local
maxima. A variation of the EM algorithm (Wright and Kennedy, 2000)
uses interval
analysis methods to locate multiple stationary points of a log
likelihood within any designated region of the parameter space.
Here, we
illustrate different ways of specification of initial value within
mixture models framework.
For independent data in the case of mixture models of ![]() components, the effect of
the E-step is to update the posterior probabilities of component
membership. Hence the first E-step can be performed
by specifying a value
components, the effect of
the E-step is to update the posterior probabilities of component
membership. Hence the first E-step can be performed
by specifying a value
![]() for each
for each
![]() , where
, where
![]() is the vector containing the
is the vector containing the ![]() posterior probabilities of
component membership for
posterior probabilities of
component membership for
![]() . The latter is usually undertaken
by setting
. The latter is usually undertaken
by setting
![]() , where
, where
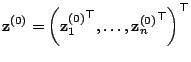 |
Another way of specifying an initial partition
![]() of
the data is to randomly divide the data into
of
the data is to randomly divide the data into ![]() groups corresponding
to the
groups corresponding
to the ![]() components of the mixture model. With random starts, the
effect of the central limit theorem tends to have the component
parameters initially being similar at least in large samples. One way
to reduce this effect is to first select a small random subsample from
the data, which is then randomly assigned to the
components of the mixture model. With random starts, the
effect of the central limit theorem tends to have the component
parameters initially being similar at least in large samples. One way
to reduce this effect is to first select a small random subsample from
the data, which is then randomly assigned to the ![]() components. The
first M-step is then performed on the basis of the subsample. The
subsample has to be sufficiently large to ensure that the first M-step
is able to produce a nondegenerate estimate of the parameter vector
components. The
first M-step is then performed on the basis of the subsample. The
subsample has to be sufficiently large to ensure that the first M-step
is able to produce a nondegenerate estimate of the parameter vector
![]() (McLachlan and Peel, 2000, Sect. 2.12). In the context
of
(McLachlan and Peel, 2000, Sect. 2.12). In the context
of ![]() normal components, a method of specifying a random start is to
generate the means
normal components, a method of specifying a random start is to
generate the means
![]() independently as
independently as
Ueda and Nakano (1998) considered a deterministic
annealing EM (DAEM) algorithm in order for the EM iterative process
to be able to recover from a poor choice of starting value. They
proposed using the principle of maximum entropy and the statistical
mechanics analogy, whereby a parameter, say ![]() , is introduced
with
, is introduced
with ![]() corresponding to the ''temperature'' in an
annealing sense. With their DAEM algorithm, the E-step is effected
by averaging
corresponding to the ''temperature'' in an
annealing sense. With their DAEM algorithm, the E-step is effected
by averaging
![]() over the distribution
taken to be proportional to that of the current estimate of the
conditonal density of the complete data (given the observed data)
raised to the power of
over the distribution
taken to be proportional to that of the current estimate of the
conditonal density of the complete data (given the observed data)
raised to the power of ![]() ; see for example
McLachlan and Peel (2000, pp. 58-60).
; see for example
McLachlan and Peel (2000, pp. 58-60).
Several methods have been suggested in the EM
literature for augmenting the EM computation with some
computation for obtaining an estimate of the covariance matrix of
the computed MLE. Many such methods
attempt to exploit the computations in the EM steps. These
methods are based on the observed information matrix
![]() , the expected information matrix
, the expected information matrix
![]() or on resampling methods. Baker (1992)
reviews such methods and also develops a method for
computing the observed information matrix in the case of
categorical data. Jamshidian and Jennrich (2000) review more
recent methods including the Supplemented EM (SEM) algorithm of
Meng and Rubin (1991) and suggest some newer methods based on
numerical differentiation.
or on resampling methods. Baker (1992)
reviews such methods and also develops a method for
computing the observed information matrix in the case of
categorical data. Jamshidian and Jennrich (2000) review more
recent methods including the Supplemented EM (SEM) algorithm of
Meng and Rubin (1991) and suggest some newer methods based on
numerical differentiation.
Theorectically one may compute the asymptotic covariance matrix by
inverting the observed or expected information matrix at the MLE.
In practice, however, this may be tedious analytically or
computationally, defeating one of the advantages of the EM
approach. Louis (1982) extracts the observed information matrix
in terms of the conditional moments of the gradient and curvature of
the complete-data log likelihood function introduced within the EM
framework. These conditional moments are generally easier to work out
than the corresponding derivatives of the incomplete-data log
likelihood function.
An alternative approach is to numerically
differentiate the likelihood function to
obtain the Hessian. In a EM-aided differentiation approach,
Meilijson (1989) suggests perturbation of the incomplete-data
score vector to compute the observed information matrix.
In the SEM algorithm (Meng and Rubin, 1991), numerical
techniques are used to compute
the derivative of the EM operator
![]() to obtain
the observed information matrix.
The basic idea is to use the fact that the rate of
convergence is governed by the fraction of the missing information
to find the increased variability due to missing information to add
to the assessed complete-data covariance matrix.
More specifically, let
to obtain
the observed information matrix.
The basic idea is to use the fact that the rate of
convergence is governed by the fraction of the missing information
to find the increased variability due to missing information to add
to the assessed complete-data covariance matrix.
More specifically, let
![]() denote the asymptotic covariance
matrix of the MLE
denote the asymptotic covariance
matrix of the MLE
![]() . Meng and Rubin (1991) show that
. Meng and Rubin (1991) show that
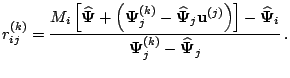 |
It is important to emphasize that estimates of the covariance
matrix of the MLE based on the expected or observed information
matrices are guaranteed to be valid inferentially only
asymptotically. In particular for mixture models, it is well known
that the sample size ![]() has to be very large before the asymptotic
theory of maximum likelihood applies. A resampling approach, the
bootstrap (Efron, 1979; Efron and Tibshirani, 1993),
has been considered to tackle this problem.
Basford et al. (1997) compared the bootstrap and
information-based approaches for some normal mixture models and
found that unless the sample size was very large, the standard
errors obtained by an information-based approach were too unstable
to be recommended.
has to be very large before the asymptotic
theory of maximum likelihood applies. A resampling approach, the
bootstrap (Efron, 1979; Efron and Tibshirani, 1993),
has been considered to tackle this problem.
Basford et al. (1997) compared the bootstrap and
information-based approaches for some normal mixture models and
found that unless the sample size was very large, the standard
errors obtained by an information-based approach were too unstable
to be recommended.
The bootstrap is a powerful technique that permits the variability in
a random quantity to be assessed using just the data at hand. Standard
error estimation of
![]() may be implemented
according to the bootstrap as follows. Further discussion on
bootstrap and resampling methods can be found in
Chaps. III.2 and
III.3 of this handbook.
may be implemented
according to the bootstrap as follows. Further discussion on
bootstrap and resampling methods can be found in
Chaps. III.2 and
III.3 of this handbook.
In Step 1 above, the nonparametric version of the bootstrap would take
![]() to be the empirical distribution function formed from
the observed data
to be the empirical distribution function formed from
the observed data
![]() . Situations where we may wish to use
the latter include problems where the observed data are censored or
are missing in the conventional sense. In these cases the use of the
nonparametric bootstrap avoids having to postulate a suitable model
for the underlying mechanism that controls the censorship or the
absence of the data. A generalization of the nonparametric version of
the bootstrap, known as the weighted bootstrap, has been studied by
Newton and Raftery (1994).
. Situations where we may wish to use
the latter include problems where the observed data are censored or
are missing in the conventional sense. In these cases the use of the
nonparametric bootstrap avoids having to postulate a suitable model
for the underlying mechanism that controls the censorship or the
absence of the data. A generalization of the nonparametric version of
the bootstrap, known as the weighted bootstrap, has been studied by
Newton and Raftery (1994).